Prenatal visits are generally happy occasions, but many women still experience some anxiety as every pregnancy comes with a risk of chromosomal abnormalities. Fortunately, advances in non-invasive prenatal test technologies (NIPT) make the testing process more convenient and reliable than ever before.
The Rise of cfDNA Analysis for NIPT
Since its introduction in the 1990s, the first trimester combined screening test (FTS) has been a common approach to prenatal testing. This approach relies on a combination of serum biochemistry and foetal nuchal translucency, typically between 11 and 13 weeks gestation. It reliably screens [1] for common chromosomal abnormalities such as Down syndrome (trisomy 21) and Edward syndrome (trisomy 18).
FTS has been a great asset but it comes with a high false positive and low positive predictive value, according to Dr Tachjaree Panchalee, a physician in the Department of Obstetrics & Gynaecology at Siriraj Hospital in Thailand, during the 2018 NIPT Forum Asia. FTS has been shown to achieve a detection rate of 84-90% with a false positive rate of 5% [2], which is better than nothing but definitely sub-optimal.
Since 2011, however, new methods have emerged to challenge FTS. These methods non-invasively screen a fetus for chromosomal abnormalities based on cell-free DNA (cfDNA) analysis.
“[FTS] has been superseded by cell-free DNA testing,” said Professor Liona Poon, a Professor in the Department of Obstetrics & Gynaecology at the Chinese University of Hong Kong, during the same event.
Fragments of placenta DNA comprise on average 13% [3] of the total cfDNA in maternal blood. Foetal DNA fragments of between 150 to 200 base pairs [4] can be found in the maternal blood from as early as day 32 of gestation and increase with time [5]. These fragments vanish from the maternal blood from as early as two-hours post delivery [3].
Analysing maternal and fetal cfDNA allows laboratory medicine experts to non-invasively and accurately assess the fetus for chromosomal abnormalities.
Laboratories that are well-versed in cfDNA analysis will be at the forefront of new applications, and will be able to add value to their partner obstetricians ahead of the curve. With access to growing validated reference libraries, the application of cfDNA testing can potentially be extended to other chromosomal abnormalities in future.
“It is the healthcare provider’s responsibility to stay up to date with available screening tools because every woman has the right to make her own choice,” added Dr Panchalee.
Approaches to cfDNA
As with any new discovery, a number of methods for NIPT are jostling for gold standard status. These range from whole genome and specific chromosome sequencing to microassay-based platforms.
One of the earliest technologies used is massively parallel (“shotgun”) sequencing. This method is not targeted to regions of interest, but rather assesses the whole genome of all cfDNA fragments present in the maternal plasma.
In whole genome sequencing [6], tens of millions of sequence reads are aligned and mapped against a human reference library to match the data obtained to the chromosome of origin. The mapped data is then counted to determine whether the fetus carries a chromosomal abnormality.
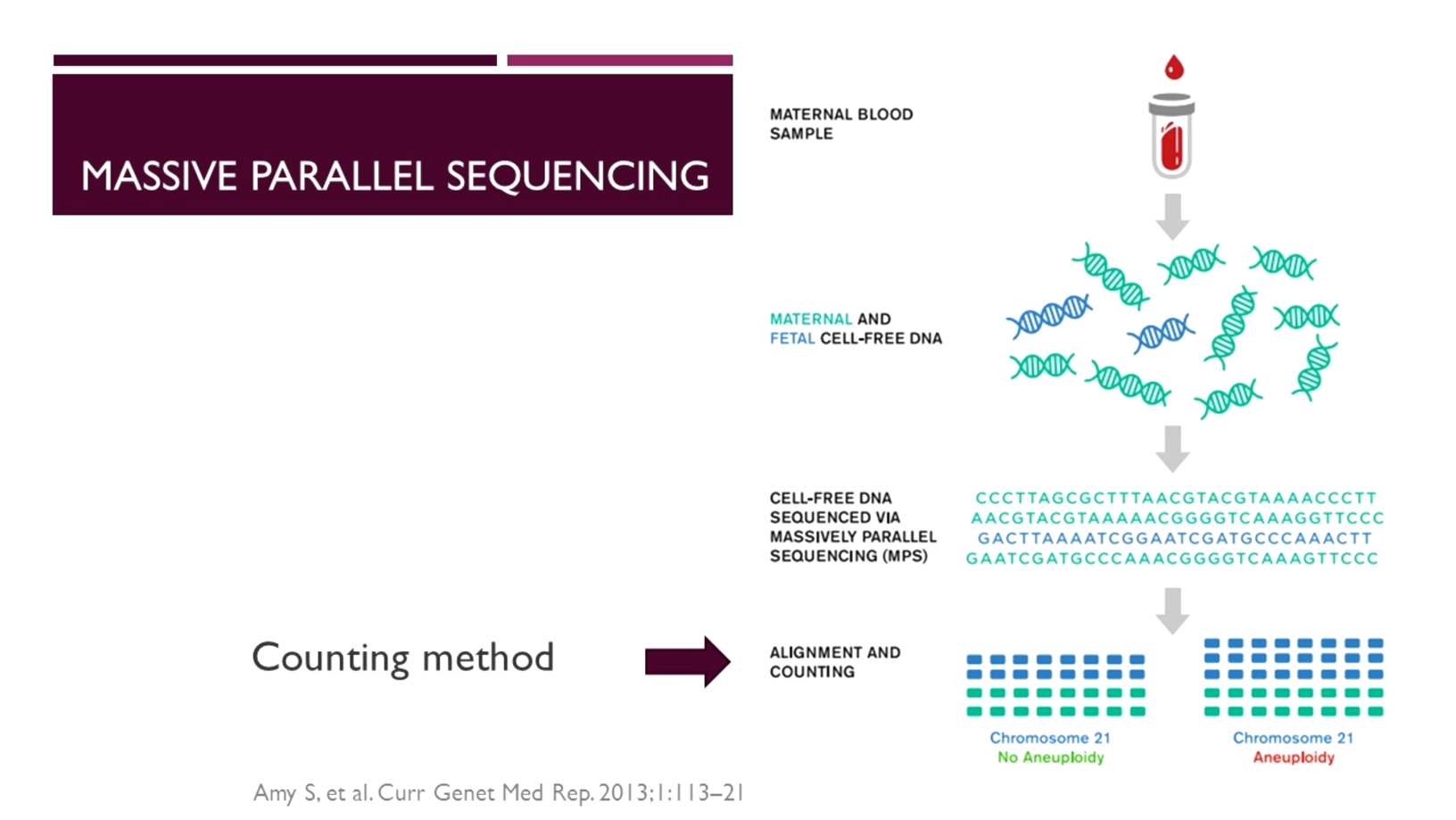
“With massively parallel sequencing, approximately 25 million raw sequencing lists [7] are required per sample to generate sufficient data for accurate analysis of the regions of interest,” said Dr Panchalee. “This approach represents enormous redundancy given that the clinically significant chromosomes represent only 14% of the genome [7].”
Newer technologies are emerging, including semiconductor sequencing technologies such as ion torrent platforms. While a number of platforms exist, in a comparison of two such technologies, researchers have found that [8] the amount of sequencing and the application of common and robust statistical analyses are more important to ensuring accurate results than the specific sequencing chemistry or platform.
More recently, microarray analysis has been adapted for cfDNA testing. With this, selected regions can be digitally analysed for specific chromosomal abnormalities, including trisomies 21, 18, 13 as well as sex chromosome aneuploidies.

In a head-to-head analysis with next generation sequencing (NGS), microarray analysis has been found to result in faster and more precise [9] cfDNA analysis than NGS. Each sample is analysed in a singleplex in each sub-array, creating a simple and cost-effective process to analyse samples. This method has been extensively validated in prospective clinical trials across various obstetric populations.
“An internal validation of the system demonstrated high accuracy and specificity, with no false positives or false negatives,” confirmed Dr Panchalee.
The latest technology to emerge is a six-step process that images single DNA molecules [10], reducing cfDNA assay complexity. It combines novel molecular probe technology to label target chromosomes with a new readout format using a nanofilter. This allows enrichment of single molecules for imaging and counting without DNA amplification microarray or sequencing. To date, this technology has not been widely validated in prospective clinical studies, unlike NGS or microarray.
What is cfDNA’s potential impact?
As a screening test offered only in high or intermediate risk pregnancies, cfDNA testing is already changing women’s choices and reducing the procedure related to miscarriage risk and anxiety. A study of pregnant women in Hong Kong [11] found that rates of invasive testing have dropped significantly following the introduction of cfDNA testing.
Laboratories that can offer obstetricians another non-invasive test method will empower pregnant women to make informed decisions more decisively and ultimately avoid unnecessary invasive procedures.
[1] Malone, D.F., et al., (2005). First-Trimester or Second-Trimester Screening, or Both, for Down’s Syndrome. The New England Journal of Medicine. 353(19), pp.2001-2011.
[2] Li, S. W., et al., (2015). The assessment of combined first trimester screening in women of advanced maternal age in an Asian cohort. Singapore medical journal, 56(1), pp.47–52.
[3] Kotsopoulou, I., et al., (2015). Non-invasive prenatal testing (NIPT): limitations on the way to become diagnosis, Diagnosis, 2(3), pp.141-158.
[4] Taglauer, E. S., et al., (2014). Review: cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta, 35 Suppl(Suppl), S64–S68.
[5] Wataganara, T., et al., (2004). Cell-free fetal DNA levels in maternal plasma after elective first-trimester termination of pregnancy. Fertility and Sterility, 81(3), pp.638-644.
[6] Swanson, A., et al., (2013). Non-invasive Prenatal Testing: Technologies, Clinical Assays and Implementation Strategies for Women’s Healthcare Practitioners. Current genetic medicine reports, 1(2), pp.113–121.
[7] Norwitz, E. R., & Levy, B. (2013). Noninvasive prenatal testing: the future is now. Reviews in obstetrics & gynecology, 6(2), pp.48–62.
[8] Kim, S., et L., (2016). Comparison of two high-throughput semiconductor chip sequencing platforms in noninvasive prenatal testing for Down syndrome in early pregnancy. BMC medical genomics, 9(1), pp.22.
[9] Juneau, K., et al., (2014). Microarray-Based Cell-Free DNA Analysis Improved Noninvasive Prenatal Testing. Fetal Diagnosis and Therapy, 36, pp.282-286.
[10] Dahl, F., et al., (2018). Imaging single DNA molecules for high precision NIPT. Scientific Reports, 8, 4549.
[11] Cheng YKY., eet al., (2018). Women’s preference for non-invasive prenatal DNA testing versus chromosomal microarray after screening for Down syndrome: a prospective study. BJOG, 125, pp.451–459.
This article is based on the presentations “Evolution of clinically relevant NIPT technology” and “Updated evidence and implementation models of cfDNA testing” at 2018 NIPT Forum Asia in Bangkok, Thailand.









